POMS: Difference between revisions
| Line 34: | Line 34: | ||
====log files==== | ====log files==== | ||
POMS will steer you to a submission log page where you can get the DAG top-level condor cluster number. Following are examples of logf iles you can see on the web or downlooad: | POMS will steer you to a submission log page where you can get the jobid which contains DAG top-level condor cluster number. Following are examples of logf iles you can see on the web or downlooad: | ||
jobsub_fetchlog --jobid=43653405.0@jobsub03.fnal.gov | jobsub_fetchlog --jobid=43653405.0@jobsub03.fnal.gov | ||
Some places to find condor logs (including evictions, holds, etc. | Some places to find condor logs (including evictions, holds, etc. | ||
Revision as of 21:04, 30 January 2023
Introduction
The Production Operations Management Service (POMS) is a computing division tool that helps users to run large and complex grid campaigns. It is designed to assist experiments in their MonteCarlo production and data processing, both for production and analysis
POMS provides
- control scripts and easy-to-use GUI interface. It is fully configurable with any executable;
- automatic monitoring and campaign management options;
- chaining of multiple stages of a grid campaign i.e. automated submission of the following stages;
- automated recovery options and easy re-submission of failed jobs;
- analysis of logs and database entries for results.
POMS has been successfully employed for the production of the MDC2020 datasets. The supported and recommended way to access POMS is through the web interface. project-py is a Python script that provides a command line interface.
The POMS system is designed around the concept of a campaign. A campaign is a set of stages which can have interdependencies. Each stage corresponds to the submission of a certain number of jobs to the computing grid, with a specific configuration. Each stage typically takes as input an entire SAM dataset and produces one or more SAM datasets as output, which contain the output files produced by each job.
Experimenters only need one or more executable files that write and/or read data files. They can then build (or borrow from an example) an .ini file for fife_launch that describes what setup scripts to run, how their executable should be run, where output files should be put, and similar details, for each stage of their processing, and define a SAM dataset of files they would like processed.
Archiecture
POMS interfaces with the existing Fermilab data access, processing, movement and monitoring tools at Fermilab, e.g. Jobsub (for job submission), SAM (for data management), PostgreSQL from the central database team and Landscape (for monitoring). In combination with these tools, POMS handles the creation of intermediate datasets of files for multistage campaigns.
The underlying batch system being used is Condor with GlideinWMS. POMS launches condor clusters or DAGs (directed acyclic graph) of jobs using fife_launch wrappers around the jobsub submission system.
POMS uses the Landscape monitoring suite via an interface called Lens, which provides summaries of the jobs in a submission so that POMS can determine when submissions are complete etc.
POMS uses SAM and its "project" functionality and provenance information to build a "recovery dataset" for the SAM project that delivered files to the submission.
See:
- EPJ Web of Conferences 245, 03024 (2020) for more on POMS
- https://dune.github.io/computing-training-202105/aio/index.html For overview of terminlogy
log files
POMS will steer you to a submission log page where you can get the jobid which contains DAG top-level condor cluster number. Following are examples of logf iles you can see on the web or downlooad:
jobsub_fetchlog --jobid=43653405.0@jobsub03.fnal.gov
Some places to find condor logs (including evictions, holds, etc.
43653405 - top level cluster, some of the untarred files: dagbegin-fife_wrap_20230128_183253_3465810.log - logging condor 43653406 - transfers input files - transfers output files?? fife_wrap_20230128_183253_3465810_1.dag.dagman.log - logging condor 43653405 - does not have individual node info fife_wrap_20230128_183253_3465810_1_1_.log - logging condor 43653409 - contains actual job work, include memory and time updates fife_wrap_20230128_183253_3465810_1.dag.nodes.log - running condor 43653406 and summary of 43653409 dagend-fife_wrap_20230128_183253_3465810.log - logging condor 43729756
Tutorial
Work in progress
In this tutorial we will create a campaign with two stages, whose goal is to run the reconstruction stage on a pre-existing digi sample. This campaign can be then easily extended to run an arbitrary number of stages (e.g. generation, digitization, and reconstruction).
The first step is to create a proxy certification that will be then used by POMS:
setup fife_utils
kx509 -n --minhours 168 -o /tmp/x509up_voms_mu2e_Analysis_${USER}
upload_file /tmp/x509up_voms_mu2e_Analysis_${USER}
A POMS campaign requires two files: a INI file, which defines the stage names and the interdependencies, and a CFG file, which describes the configuration for each stage.
INI file
Let's start with the INI file: it can be created locally and it is then uploaded to POMS through the web interface. In the following INI file we will specify two stages: the first one creates the FCL files, one per job, and the second one takes as input the SAM dataset containing the FCL files and submit N jobs, where N is the number of FCL files in the dataset.
First of all, the INI file needs the definition of the campaign and of the job type we are going to run.
[campaign] experiment = mu2e poms_role = analysis name = srsoleti_tutorial campaign_stage_list = reco_fcl, reco [campaign_defaults] vo_role=Analysis software_version=MDC2020t dataset_or_split_data=None cs_split_type=None completion_type=complete completion_pct=100 param_overrides="[]" test_param_overrides="[]" merge_overrides=False login_setup=srsoleti_poms_login job_type=mu2e_reco_srsoleti_jobtype stage_type=regular output_ancestor_depth=1 [login_setup srsoleti_poms_login] host=pomsgpvm01.fnal.gov account=poms_launcher setup=setup fife_utils v3_5_0, poms_client, poms_jobsub_wrapper;
Then, we define two job types, one that corresponds to the stage that will run the mu2e process, and one that corresponds to the stage that will run the generate_fcl script, which creates the FCL files.
[job_type mu2e_reco_srsoleti_jobtype] launch_script = fife_launch parameters = [["-c ", "/mu2e/app/users/srsoleti/tutorial/reco.cfg"]] output_file_patterns = %.art recoveries = [["proj_status",[["-Osubmit.dataset=","%(dataset)s"]]]] [job_type generate_fcl_reco_srsoleti_jobtype] launch_script = fife_launch parameters = [["-c ", "/mu2e/app/users/srsoleti/tutorial/reco.cfg"]] output_file_patterns = %.fcl
Finally, we define the two stages that form our campaign, reco and reco_fcl.
[campaign_stage reco_fcl] param_overrides = [["--stage ", "reco_fcl"]] test_param_overrides = [["--stage ", "reco_fcl"]] job_type = generate_fcl_reco_srsoleti_jobtype [campaign_stage reco] param_overrides = [["--stage ", "reco"]] test_param_overrides = [["--stage ", "reco"]] [dependencies reco] campaign_stage_1 = reco_fcl file_pattern_1 = %.fcl
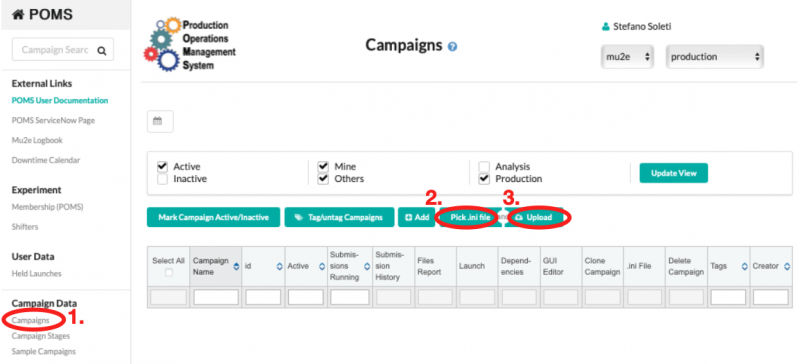
The file can then uploaded to POMS by connecting to POMS, clicking on "Campaigns", then "Pick .ini file", and "Upload", as shown in the picture below.
CFG file
The CFG file describes the configuration for each stage and it will essentially tell the grid node what to run and in which order. First, we start with a [global] where we define variables that will be used later in the file
[global]
group = mu2e
subgroup = highpro
experiment = mu2e
wrapper = file:///${FIFE_UTILS_DIR}/libexec/fife_wrap
submitter = srsoleti
outdir_sim = /pnfs/mu2e/tape/usr-sim/sim/%(submitter)s/
outdir_dts = /pnfs/mu2e/tape/usr-sim/dts/%(submitter)s/
logdir_bck = /pnfs/mu2e/tape/usr-etc/bck/%(submitter)s/
outdir_mcs = /pnfs/mu2e/tape/usr-sim/mcs/%(submitter)s/
primary_name = CeEndpoint
stage_name = override_me
artRoot_dataset = override_me
histRoot_dataset = override_me
override_dataset = override_me
release = MDC2020
release_v_i = r
release_v_o = t
desc = %(release)s%(release_v_o)s
db_folder = mdc2020t
db_version = v1_0
db_purpose = perfect
beam = 1BB
As you can see this is quite verbose. However, future CFG files can be made slimmer by using the includes statement in the [global] section, which import existing CFG files.
Now, we need to define the default job configuration. This can also be written in a separate CFG file and imported.
[submit]
debug = True
G = %(group)s
subgroup = %(subgroup)s
e = SAM_EXPERIMENT
e_1 = IFDH_DEBUG
e_2 = POMS4_CAMPAIGN_NAME
e_3 = POMS4_CAMPAIGN_STAGE_NAME
resource-provides = usage_model=DEDICATED,OPPORTUNISTIC
generate-email-summary = True
expected-lifetime = 23h
memory = 2500MB
email-to = %(submitter)s@fnal.gov
l = '+SingularityImage=\"/cvmfs/singularity.opensciencegrid.org/fermilab/fnal-wn-sl7:latest\"'
append_condor_requirements='(TARGET.HAS_SINGULARITY=?=true)'
f = dropbox:///mu2e/app/home/mu2epro/db_fcl_test/ucondb_auth.sh.tar.gz
[job_setup]
debug = True
find_setups = False
source_1 = /cvmfs/mu2e.opensciencegrid.org/setupmu2e-art.sh
source_2 = /cvmfs/mu2e.opensciencegrid.org/Musings/SimJob/%(release)s%(release_v_o)s/setup.sh
source_3 = $CONDOR_DIR_INPUT/ucondb_auth.sh
setup_1 = -B ifdh_art v2_14_06 -q e20:prof
setup_2 = -B mu2etools
setup_3 = -B sam_web_client
ifdh_art = True
postscript = find *[0-9]* -maxdepth 1 -name "*.fcl" -exec sed -i "s/MU2EGRIDDSOWNER/%(submitter)s/g" {} +
postscript_2 = find *[0-9]* -maxdepth 1 -name "*.fcl" -exec sed -i "s/MU2EGRIDDSCONF/%(desc)s/g" {} +
postscript_3 = find *[0-9]* -maxdepth 1 -name "*.fcl*" -exec mv -t . {} +
postscript_4 = [ -f template.fcl ] && rm template.fcl
postscript_5 = [[ $(ls *.art) ]] && samweb file-lineage parents `basename ${fname}` > parents.txt
postscript_6 = [[ $(ls *.art) ]] && echo ${fname} >> parents.txt
[sam_consumer]
limit = 1
schema = xroot
appvers = %(release)s
appfamily = art
appname = SimJob
[executable]
name = loggedMu2e.sh
Now, we have to define the kind of output files (the "job outputs") created by our jobs. They are three: the .fcl files generated by the dedicated stage, the .tbz files containing the log of the mu2e process, and the .art files created by the mu2e process.
[job_output] addoutput = cnf.*.fcl add_to_dataset = cnf.%(submitter)s.%(stage_name)s.%(desc)s.fcl declare_metadata = True metadata_extractor = json add_location = True [job_output_1] addoutput = *.tbz declare_metadata = False metadata_extractor = printJsonSave.sh add_location = True add_to_dataset = bck.%(submitter)s.%(stage_name)s.%(desc)s.tbz hash = 2 hash_alg = sha256 [job_output_2] addoutput = *.art declare_metadata = True metadata_extractor = printJsonSave.sh add_location = True hash = 2 hash_alg = sha256
The final step is to define the two stages: one called reco_fcl which submits 1 job to the grid, whose goal is to create, save, and declare the FCL files, and one called reco which submits one job for each FCL file present in the input SAM dataset.
[stage_reco_fcl] global.stage_name = %(primary_name)sMix%(beam)sSignal10 global.desc = %(release)s%(release_v_o)s_%(db_purpose)s_%(db_version)s job_setup.ifdh_art = False job_output.dest = https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/data/%(db_folder)s/ executable.name = gen_Reco.sh executable.arg_1 = %(stage_name)s executable.arg_2 = %(release)s executable.arg_3 = %(release_v_i)s executable.arg_4 = %(release_v_o)s executable.arg_5 = %(db_purpose)s executable.arg_6 = %(db_version)s executable.arg_7 = 1 executable.arg_8 = %(submitter)s [stage_reco] global.stage_name = %(primary_name)sMix%(beam)sSignal global.desc = %(release)s%(release_v_o)s_%(db_purpose)s_%(db_version)s global.artRoot_dataset = mcs.%(submitter)s.%(stage_name)s.%(desc)s.art job_output_1.dest = %(logdir_bck)s/%(stage_name)s/%(desc)s/tbz/ job_output_2.addoutput = mcs.*.%(stage_name)s.*art job_output_2.dest = %(outdir_mcs)s/%(stage_name)s/%(desc)s/art submit.dataset = cnf.%(submitter)s.%(stage_name)s10.%(desc)s.fcl submit.n_files_per_job = 1 sam_consumer.limit = 1 sam_consumer.schema = https job_setup.getconfig = True
The gen_Reco.sh, whose source can be found here, generates the FCL files needed for the reconstruction step. The reco stage then takes as input those FCL files and run the corresponding jobs.
Submitting the campaign
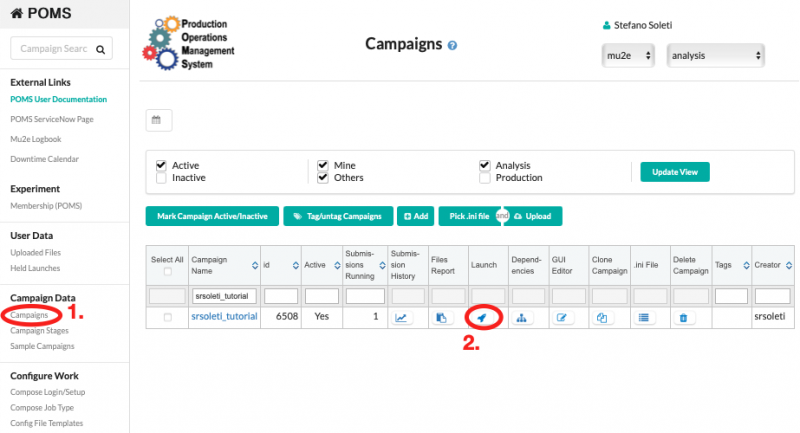
Once the INI file has been uploaded, it is possible to submit the campaign on the POMS web interface, as shown in the screenshot below. The system will submit the reco_fcl stage and, once that is completed, it will automatically submit the reco one.
UconDB
mu2e_ucon_prod was created 11/2021 to help with MDC2020. general ucondb docs and more docs
- database mu2e_ucon_prod owned by nologin role mu2e_ucon_prod;
- Kerberos-authenticated roles: brownd kutschke srsoleti
- md5 authenticated role 'mu2e_ucon_web' (for POMS)
- port is 5458 (on ifdbprod/ifdb08)
https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/... - not cached, external and internal access http://dbdata0vm.fnal.gov:9090/mu2e_ucondb_prod/app/... - not cached, internal access only https://dbdata0vm.fnal.gov:8444/mu2e_ucondb_prod/app/... - cached, external and internal access http://dbdata0vm.fnal.gov:9091/mu2e_ucondb_prod/app/... - cached, internal access only
The following curl command create a folder in the database called "test":
curl --digest -u mu2e:$DB_PWD https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/create_folder?folder=test
and to insert a file
curl -T <file.fhicl> —digest -u mu2e:<password> https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/data/<folder name>/<file.fhicl>
The password $DB_PWD can be requested from the Production manager.
Files in a folder can be listed, formatted, by links:
https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/UI/index
https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/UI/folder?folder=mdc2020r
or as text at the command line:
curl "https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/folders" curl "https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/objects?folder=mdc2020r"
and a fcl file can be retrieved:
curl "https://dbdata0vm.fnal.gov:9443/mu2e_ucondb_prod/app/data/mdc2020r/cnf.mu2e.CeEndpointMix2BB.MDC2020r_perfect_v1_0.001210_00000771.fcl"
The POMS FCL stages take care of saving the FCL files. The SAM location of the files stored in the database looks like this:
$ samweb locate-file cnf.mu2e.POT.db_test_v12.001201_00000001.fcl dbdata0vm.fnal.gov:/mu2e_ucondb_prod/app/data/cnf_mu2e_POT_db_test_v12_fcl
Updating SAM Records
Updating SAM records with tape locations is a standard part of POMS based workflow. Tape location is generally added automatically. There is a program that is run twice a day to read the DCache transfer logs and update SAM with the tape location information for files copied to tape.
However, this read-the-dcache-logs approach does sometimes miss things, often due to files missing in the boundary between logs, or if the log disk on the dCache servers is run short of space, etc.
In this case the solution is to do periodically something like this:
export SAM_EXPERIMENT=mu2e samweb create-definition missing_tape_2021_07_22 " (start_time > '2021-07-01T00:00:00' and start_time < '2021-07-20T00:00:00' and full_path like '/pnfs/$SAM_EXPERIMENT/tape/%' ) minus tape_label like '%'" samweb count-files defname:missing_tape_2021_07_22 sam_validate_dataset --tapeloc --name missing_tape_2021_07_22 samweb list-files defname:missing_tape_2021_07_22 > files_not_on_tape
References
- POMS production top
- redmine
- DUNE video tutorial